Proteins are fundamental building blocks of life, responsible for a wide range of biological functions. They are involved in catalyzing chemical reactions, transmitting signals between cells, and providing structural support for cells and tissues. However, in order to perform these functions, proteins must first assume a specific three-dimensional shape or conformation, known as their native state. The process by which proteins adopt their native state is known as protein folding.
Protein folding is a complex process that occurs spontaneously in living cells. It involves the formation of intricate and highly specific patterns of intermolecular interactions that dictate the protein’s final shape. These interactions are primarily driven by the physical properties of the amino acids that make up the protein sequence, including their size, shape, charge, and hydrophobicity.
The importance of protein folding is underscored by the fact that when proteins fail to fold correctly, they can lead to a variety of diseases. These include Alzheimer’s, Parkinson’s, and cystic fibrosis, among others. In fact, a significant portion of the pharmaceutical industry is focused on developing drugs that can target misfolded proteins and restore their proper function.
Understanding protein folding is therefore a critical area of study in molecular biology and biochemistry.
The Basics of Protein Folding
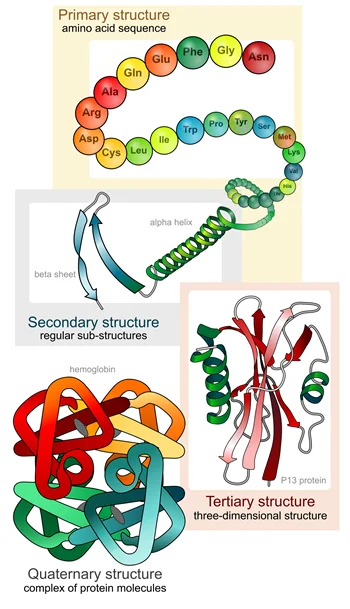
Protein folding occurs in several stages, beginning with the formation of the primary structure, which is simply the linear sequence of amino acids that make up the protein. The amino acid sequence is determined by the genetic code, which specifies the order of the nucleotides in DNA.
Once the primary structure is formed, the protein begins to fold into its native conformation. The first step in this process is the formation of local structural elements, known as secondary structures. These include alpha-helices, beta-sheets, and turns. These structures are stabilized by hydrogen bonds between the amino acid side chains, as well as other intermolecular interactions.
The next stage in protein folding is the formation of tertiary structure, which involves the folding of the secondary structures into a compact, three-dimensional shape. This is often driven by hydrophobic interactions, as nonpolar amino acids cluster together to minimize contact with water.
Finally, in some cases, multiple protein chains can come together to form a quaternary structure, which further enhances the protein’s stability and function.
Factors that Influence Protein Folding
The folding process is influenced by a variety of factors, including the physical properties of the amino acids, the environment in which the protein is folding, and the presence of chaperone proteins that assist in the folding process.
One important factor is the amino acid sequence itself. Certain amino acids, such as proline, have rigid structures that can limit the flexibility of the protein backbone, making it more difficult to fold. Other amino acids, such as cysteine, can form covalent bonds with each other, leading to the formation of disulfide bridges that can stabilize the protein’s structure.
The environment in which the protein is folding can also have a significant impact. For example, the presence of denaturing agents, such as urea or guanidinium chloride, can disrupt the protein’s structure and prevent proper folding. Conversely, the presence of molecular chaperones, specialized proteins that assist in the folding process, can enhance the protein’s folding and stability.
Studying protein folding is a challenging endeavor, given the complexity of the process and the difficulty in visualizing the three-dimensional structure of proteins. However, a variety of techniques have been developed to probe the folding process and gain insights into its underlying mechanisms.
- Circular Dichroism Spectroscopy (CD)
Circular dichroism spectroscopy is a technique that measures the difference in the absorption of left- and right-circularly polarized light by chiral molecules such as proteins. This difference in absorption arises due to the asymmetric structure of the protein molecule and is sensitive to changes in the protein’s secondary structure. By measuring the changes in the CD signal over time as a protein is folded, researchers can gain insights into the structural changes that occur during folding.
- Fluorescence Spectroscopy
Fluorescence spectroscopy is a powerful technique for studying protein folding, as it can monitor changes in the fluorescent properties of amino acids as they undergo conformational changes. This technique is often used to monitor the exposure of hydrophobic residues, which are typically buried within the protein’s interior in the folded state. As the protein unfolds, these residues become exposed to water, leading to changes in their fluorescent properties that can be monitored by spectroscopy.
- Nuclear Magnetic Resonance (NMR) Spectroscopy
Nuclear magnetic resonance spectroscopy is a technique that is widely used to study protein structure and dynamics. NMR can be used to determine the three-dimensional structure of proteins in solution, providing valuable insights into the folding process. In addition, NMR can be used to study protein dynamics, allowing researchers to monitor changes in the protein’s conformation over time.
- X-ray Crystallography
X-ray crystallography is a powerful technique for determining the three-dimensional structure of proteins. This technique involves growing crystals of the protein of interest and then exposing the crystals to X-rays. The X-rays interact with the atoms in the crystal, producing a diffraction pattern that can be used to determine the positions of the atoms in the protein. X-ray crystallography has been used to determine the structures of thousands of proteins, providing valuable insights into protein folding and function.
- Single-Molecule Techniques
Single-molecule techniques, such as single-molecule fluorescence resonance energy transfer (smFRET) and atomic force microscopy (AFM), allow researchers to study the folding process of individual protein molecules in real time. These techniques are particularly useful for studying the folding of large, complex proteins, which may be difficult to study using other techniques. By observing the folding process of individual molecules, researchers can gain insights into the kinetics and thermodynamics of the folding process.
There have been several real-time cases of protein folding studies that have provided insights into the mechanisms of protein folding and its importance in various biological processes. Here are a few examples:
- Protein Folding in Alzheimer’s Disease
Alzheimer’s disease is a neurodegenerative disorder that is characterized by the accumulation of amyloid beta (Aβ) peptides in the brain. These peptides are derived from the amyloid precursor protein (APP), and their aggregation into insoluble plaques is thought to be a critical factor in the development of Alzheimer’s disease. Recent studies have used single-molecule fluorescence techniques to study the folding of APP and Aβ peptides in real time. These studies have shown that Aβ peptides can exist in a range of conformational states, some of which are prone to aggregation, while others are not. Understanding the folding of Aβ peptides and the factors that influence their aggregation could lead to the development of new therapies for Alzheimer’s disease.
- Protein Folding in Cancer
Cancer is a disease that is characterized by uncontrolled cell growth and proliferation. Many cancer cells rely on specific signaling pathways to grow and survive, and these pathways are often regulated by proteins that undergo conformational changes in response to stimuli. One example is the epidermal growth factor receptor (EGFR), which is a protein that is often overexpressed in cancer cells. Recent studies have used NMR spectroscopy to study the conformational changes that occur in the EGFR protein in response to ligand binding. These studies have provided insights into the mechanisms by which EGFR regulates cell growth and survival, and could lead to the development of new therapies for cancer.
- Protein Folding in Antibody Design
Antibodies are proteins that play a critical role in the immune system, by recognizing and neutralizing foreign pathogens such as viruses and bacteria. In recent years, there has been growing interest in the design of new antibodies for therapeutic use, such as for the treatment of cancer and autoimmune diseases. One approach to antibody design involves the use of computational techniques to predict the folding of the antibody protein and to optimize its structure for specific binding properties. Real-time studies of protein folding using techniques such as NMR and X-ray crystallography have been critical for validating these computational predictions, and for understanding the factors that influence the folding and stability of antibody proteins.
- Protein Folding in Enzyme Catalysis
Enzymes are proteins that catalyze biochemical reactions in living organisms. The catalytic activity of enzymes is often regulated by conformational changes that occur during the folding of the protein. Real-time studies of enzyme folding using techniques such as NMR and X-ray crystallography have provided insights into the mechanisms of enzyme catalysis, and have led to the development of new enzyme inhibitors for the treatment of diseases such as cancer and infectious diseases.
Artificial intelligence (AI) is revolutionizing the field of protein folding by providing novel approaches to predict protein structure and function. Here are a few ways AI is useful in protein folding:
- Predicting Protein Structure
One of the primary applications of AI in protein folding is the prediction of protein structure from its amino acid sequence. AI algorithms can be trained on large datasets of known protein structures to learn patterns and relationships between the amino acid sequence and the resulting protein structure. These algorithms can then be used to predict the structure of proteins with unknown structures, which is critical for understanding protein function and developing new therapies.
- Enhancing Protein Design
AI can also be used to design new proteins with specific functions. By simulating the folding process and predicting the resulting protein structure, AI algorithms can optimize the amino acid sequence to achieve the desired protein function. This approach has applications in drug discovery, where new proteins can be designed to target specific disease pathways.
- Accelerating Drug Discovery
AI algorithms can also be used to accelerate the drug discovery process by predicting the binding of potential drug molecules to target proteins. This approach can significantly reduce the time and cost of drug development by identifying promising drug candidates before they are synthesized and tested in the lab.
- Identifying Protein Mutations
AI algorithms can also be used to identify mutations in proteins that are associated with the disease. By analyzing large datasets of protein sequences and structures, AI can identify mutations that are likely to disrupt protein folding and function. This approach can help identify new targets for drug development and lead to a better understanding of the underlying mechanisms of disease.
In conclusion, AI is transforming the field of protein folding by providing novel approaches to predict protein structure and function, designing new proteins with specific functions, accelerating drug discovery, and identifying protein mutations associated with the disease. The combination of AI and experimental techniques such as NMR spectroscopy and X-ray crystallography has the potential to revolutionize our understanding of protein folding and lead to the development of new therapies for a range of diseases.
AlphaFold developed by Google’s DeepMind, AlphaFold is a deep learning system that can predict the structure of proteins with remarkable accuracy. This has significant implications for drug discovery and other medical applications. AlphaFold is a deep learning-based algorithm developed by DeepMind that can predict the 3D structures of proteins with remarkable accuracy. The ability to accurately predict the 3D structure of proteins is essential for understanding how they function and for designing new drugs to target them.
Before the development of AlphaFold, predicting the 3D structure of a protein was a complex and time-consuming process that often required years of trial and error experiments.













