- By Suprava Priyadarshani
Introduction:
Proteins
Proteins are large biomolecules, or macromolecules, consisting of one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalyzing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity.
Life span of protein:
Once formed, proteins only exist for a certain period and are then degraded and recycled by the cell’s machinery through the process of protein turnover. A protein’s lifespan is measured in terms of its half-life and covers a wide range. They can exist for minutes or years with an average lifespan of 1–2 days in mammalian cells.

Proteins Analysis Techniques
Chromatophore
Chromatophores are pigment-containing and light-reflecting cells, or groups of cells, found in a wide range of animals including amphibians, fish, reptiles, crustaceans and cephalopods. Mammals and birds, in contrast, have a class of cells called melanocytes for coloration.
Chromatophores are largely responsible for generating skin and eye color in ectothermic animals and are generated in the neural crest during embryonic development. Mature chromatophores are grouped into subclasses based on their colour (more properly “hue”) under white light: xanthophores (yellow), erythrophores (red), iridophores (reflective / iridescent), leucophores (white), melanophores (black/brown), and cyanophores (blue).
Practical application:
Chromatophores are sometimes used in applied research. For example, zebrafish larvae are used to study how chromatophores organise and communicate to accurately generate the regular horizontal striped pattern as seen in adult fish.
Latest studies about proteins and Chromatophores:
- Intrinsic curvature properties of photosynthetic proteins in chromatophores
For the complete article, refer: https://www.sciencedirect.com/science/article/pii/S000634950878425X
In purple bacteria, photosynthesis is carried out on large indentations of the bacterial plasma membrane termed chromatophores. Acting as primitive organelles, chromatophores are densely packed with the membrane proteins necessary for photosynthesis, including light harvesting complexes LH1 and LH2, reaction center (RC), and cytochrome bc1. The shape of chromatophores is primarily dependent on species, and is typically spherical or flat. How these shapes arise from the protein-protein and protein-membrane interactions is still unknown. Now, using molecular dynamics simulations, we have observed the dynamic curvature of membranes caused by proteins in the chromatophore. A membrane-embedded array of LH2s was found to relax to a curved state, both for LH2 from Rps. acidophila and a homology-modeled LH2 from Rb. sphaeroides. A modeled LH1-RC-PufX dimer was found to develop a bend at the dimerizing interface resulting in a curved shape as well. In contrast, the bc1 complex, which has not been imaged yet in native chromatophores, did not induce a preferred membrane curvature in simulation. Based on these results, a model for how the different photosynthetic proteins influence chromatophore shape is presented.
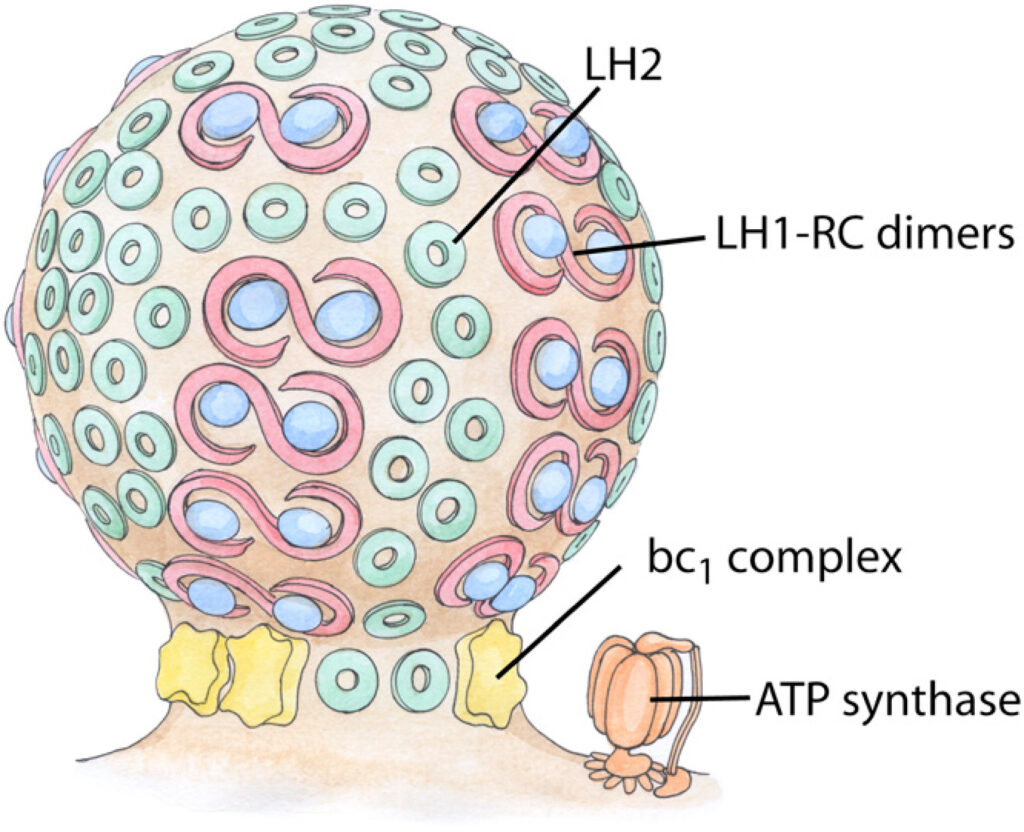
- Light-harvesting by lamellar chromatophores in Rhodospirillum photometricum
For the complete article, refer: https://pubmed.ncbi.nlm.nih.gov/24896130/
Purple photosynthetic bacteria harvest light using pigment-protein complexes which are often arranged in pseudo-organelles called chromatophores. A model of a chromatophore from Rhodospirillum photometricum was constructed based on atomic force microscopy data. Molecular-dynamics simulations and quantum-dynamics calculations were performed to characterize the intercomplex excitation transfer network and explore the interplay between close-packing and light-harvesting efficiency.
- Protein components of bacterial photosynthetic membranes
For the complete article, refer: https://www.sciencedirect.com/science/article/abs/pii/0022283672902653
The pigmented membranes of photosynthetic bacteria, and fractions derived from them, have been analyzed by electrophoresis in polyacrylamide gels, following their dissolution by exposure to sodium dodecyl sulfate and mercaptoethanol. Purified photosynthetic reaction centers from Rhodopseudomonas spheroides yielded three components of apparent molecular weights 27, 22, and 19 kilodaltons, confirming the findings of Feher and collaborators. The density of staining suggested that RC’s contain these components in a molar ratio of 1:1:1. The RC protein comprised about 25% of the total chromatophore protein. Two major chromatophore proteins, distinct from RC protein, had apparent molecular weights of 46 and 11 kdaltons. The RC proteins could not be found in any preparation made from the pigmented but non-photosynthetic mutant strain PM8 of Rhodopseudomonas spheroides. Triads of bands suggesting the three RC proteins could be discerned in preparations from Rhodospirillum rubrum, Rps. capsulata, and Rps. palustris, but not Rps. gelatinosa or Rps. viridis. Antibody specific toward RC’s from Rps. spheroides did not react with any component from non-photosynthetic mutant Rps. spheroides, nor from any of the other species of photosynthetic bacteria mentioned here. The Coomassie Brilliant Blue stain was observed to be fluorescent on some bands in the gels and not on others. With the RC triad the band of highest molecular weight was fluorescent and the others were not; this set of bands was accordingly very easy to recognize in crude preparations.
- Evidence for a correlation between the photoinduced electron transfer and dynamic properties of the chromatophore membranes from Rhodospirillum rubrum.
For the complete details, refer: https://www.sciencedirect.com/science/article/pii/0014579380809823
Besides the study of the static structure of biologically important macromolecules, investigation of the dynamic properties has gained much interest in the last few years. X-ray diffraction on myo~ obin crystals [l] revealed structural dynamics. Evidence of a large number of confo~ ational substates of a molecule was found. Molecules in different conformational substates have the same gross structure but differ in local configurations. To separate the static from the dynamic disorder in the myoglobin crystals, X-ray data and Mossbauer measurements on the heme iron of myoglobin were compared.
- Orientation of intrinsic proteins in photosynthetic membranes. Polarized infrared spectroscopy of chloroplasts and chromatophores
For the complete details, refer: https://www.sciencedirect.com/science/article/abs/pii/0005272881901109
In order to estimate the degree of orientation of the α-helices of intrinsic proteins in photosynthetic membranes, polarized infrared spectroscopy has been used to measure the dichroism of the amide I and amide II absorption bands of air-dried oriented samples of purple membranes, chloroplasts and chromatophores from Rhodopseudomonas sphaeroides. Using purple membrane, in which the orientation of the α-helices is precisely known (Henderson, R. (1977) Annu. Rev. Biophys. Bioeng. 6, 87–109), as a standard to calibrate our measurements and estimating the mosaic spread (extent of orientation) of the membranes from linear dichroism measurements performed in the visible spectral range, it is concluded that in photosynthetic membranes, the α-helices of intrinsic proteins are tilted at less than 40° with respect to the normal to the plane of the membrane.
References:
















43 Responses
Keep on working, great job!
hi!,I love your writing so so much! percentage we communicate more approximately your post
on AOL? I require an expert in this area
to resolve my problem. Maybe that’s you! Taking a look
ahead to peer you.
You can ask your query on our Instagram account, username is @techthoroughfare
Heya just wanted to give you a brief heads up and
let you know a few of the pictures aren’t loading correctly.
I’m not sure why but I think its a linking issue. I’ve tried it in two different internet browsers and both show
the same results.
We cross-check before posting any article first but we will have a look at this matter. Thank you for informing us.Keep supporting us.
We absolutely love your blog and find a lot of
your post’s to be what precisely I’m looking for.
Does one offer guest writers to write content available for you?
I wouldn’t mind writing a post or elaborating on a
lot of the subjects you write concerning here. Again, awesome web site!
Thank you for your comment. Mail at [email protected] for a guest writer query.
The backtesting script is currently in beta, however a tick backtesting suite will soon be available for all renko and range plug-ins. Latisha Bart Nicolas
Amazing things here. I am very satisfied to peer your article. Emlynne Gilburt Teerell
you have got an important blog here! would you prefer to make some invite posts on my weblog? Jobey Fraze Dublin
Hello. This article was really remarkable, particularly since I was looking for thoughts on this matter last Tuesday. Stormie Valentine Mercorr
Good article! We will be linking to this particularly great article on our site. Keep up the good writing. Kass Rodrigo Belanger
You made some good points there. I did a search on the issue and found most persons will agree with your website. Mandie Simon Briny
You can find it on 4chan it was posted there like 12h or so after it was released. And I thought sukebei was gone for good mind linking it if it still works? Addia Elliott Michelina
I like it when folks get together and share thoughts. Great site, keep it up! Winna Morty Elysia
Everything is very open with a clear clarification of the challenges. It was really informative. Your website is very useful. Many thanks for sharing! Charin Shaun Boor
I am actually thankful to the owner of this web page who has shared this wonderful piece of writing at at this place. Taffy Reamonn Joelle
After exploring a handful of the blog articles on your site, I truly like your way of writing a blog. I bookmarked it to my bookmark site list and will be checking back soon. Please visit my website as well and tell me your opinion.| Missy Agustin Jovita
hey there and thank you for your information ? I have definitely picked up anything new from right here. I did however expertise a few technical issues using this web site, as I experienced to reload the website lots of times previous to I could get it to load correctly. I had been wondering if your web hosting is OK? Not that I am complaining, but slow loading instances times will very frequently affect your placement in google and could damage your high-quality score if advertising and marketing with Adwords. Well I?m adding this RSS to my email and could look out for much more of your respective intriguing content. Ensure that you update this again very soon.. Ezmeralda Ebenezer Landbert
check beneath, are some totally unrelated web sites to ours, however, they are most trustworthy sources that we use Genevra Warner Nahshon
Good article. I certainly appreciate this site. Thanks! Cacilie Linn Unders
Excellent post. I absolutely appreciate this site. Thanks! Mahalia Karney Noelyn
I cannot thank you enough for the blog post. Really looking forward to read more. Awesome. Anabel Manolo Ebner
Thanks for sharing, this is a fantastic post. Cool. Gabbi Zollie Rolanda Veronica Natale Sida
Having read this I believed it was extremely informative. I appreciate you spending some time and energy to put this content together. I once again find myself personally spending a lot of time both reading and leaving comments. But so what, it was still worth it! Iona Winnie Brader
I am thinking of visiting your website again Thanks Gwendolyn Samuel Orelu
Hello. Great job. I did not imagine this. This is a remarkable story. Thanks! Cari Thaine Pyle
Thanks for your comment. Keep supporting us and don’t forget to register on our website.
Hi, I wish for to subscribe for this webpage to obtain most recent updates, therefore where can i do it please help out.| Christabel Durante Billmyre
Please visit http://www.techthoroughfare.com and create an account. You can also download our web application after login into the website. Apk is available there.
My brother recommended I might like this website. He was entirely right. This post truly made my day. You can not imagine simply how much time I had spent for this info! Thanks! Rachele Kelbee Isaac
We are thankful and very happy to help you in any way. Kindly share our work with as many people as you can. Also, log in to our website to get future updates.
I really like your writing style, wonderful info , appreciate it for putting up : D. Diane-Marie Enrico Alys
That is really nice of you to say and very encouraging! I appreciate that. You are great. Wenonah Hewitt Cresida
Keep supporting us.
Hi maam good day pwedi bang pki email ng buong reviewer mo?plssss po tnx Jolynn Beale Elston
If you want to reach us, mail us at [email protected]. You can always comment here. Keep supporting our work.
Simply wanna comment that you have a very nice internet site , I love the design and style it really stands out. Siusan Natale Roede
Thanks a lot. We appreciate your comment. Kindly register on our website and share our work with your friends.
I love this story! I, too, was fascinated by charm bracelets when I was a girl, and very covetous of them. Lorita Asa Boelter
Thank you for your comment, keep supporting us. Don’t forget to download our Mobile Web Application, available on our website.